Scott Lundberg
Member of Technical Staff
Microsoft AI
Biography
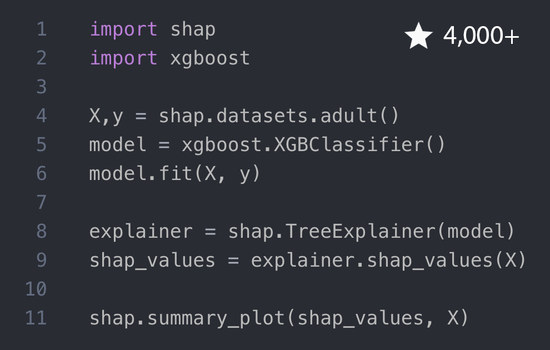
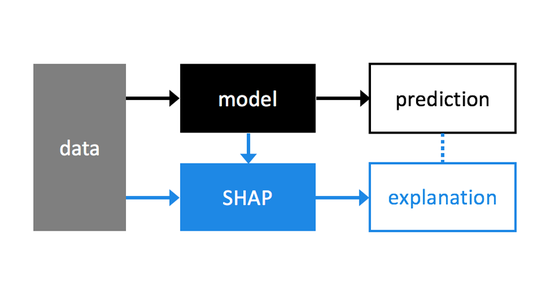
I am a Member of Technical Staff at Microsoft AI (Health), and an Affiliate Assistant Professor at the University of Washington. Previously I was a Staff Research Scientist at Google DeepMind, and a Senior Researcher at Microsoft Research. My work focuses on large language models, explainable artificial intelligence, and their application to problems in medicine and healthcare. This has led to the development of broadly applicable methods and tools for complex machine learning models that are now used in banking, logistics, manufacturing, cloud services, economics, sports, and other areas. I did my Ph.D. studies at the Paul G. Allen School of Computer Science & Engineering of the University of Washington working with Su-In Lee.
Interests
- Language Models
- Explainable AI
- Machine Learning
- Healthcare
- Genomics
Education
-
PhD in Computer Science, 2019
University of Washington
-
MS in Computer Science, 2008
Colorado State University
-
BS in Computer Science, 2005
Colorado State University